Integration of spatially resolved transcriptomics into pathological research: Opportunities and Challenges
DOI: https://doi.org/10.47184/tp.2024.01.07The development of spatially resolved transcriptomics technologies has revolutionised research in recent years. By enabling the analysis of the state and position of cell types within a tissue section, these technologies have the potential to transform our understanding of pathological processes and translate this knowledge into improved treatments for patients. This review provides an overview of available technologies and discusses the potential challenges of integrating them into pathological research, with a particular focus on the computational analysis of such data.
Keywords: Spatial transcriptomics, computational analysis, Digital Pathology, In situ sequencing, RNA sequencing
Advances in Characterizing Biological Tissues: A Focus on Spatially Resolved Transcriptomics
Since Rudolf Virchow described the cell as the central unit of life and disease in his Cellularpathologie of 1858 [1], researchers have been striving to develop new methods for characterizing the cellular composition of biological tissues. For centuries, cells have been identified by their morphology and function. In recent years, the discovery of biomolecules as the building blocks of cells has revolutionized the understanding of cell types [2] and they are now understood as manifestations of the cells’ molecular composition, adapted to their specific functions [3]. By allowing researchers to quantify different classes of biomolecules in their entirety, the emergence of omics technologies revolutionized our understanding of the molecular composition of tissues. In particular the investigation of RNA molecules, known as transcriptomics technologies has led to major breakthroughs in the identification of cell types. Technological advances have facilitated the study of the whole transcriptome of single cells and initiated consortia such as the Human Cell Atlas, which set itself the task to define all human cell types in terms of distinct molecular profiles and correlate this information with the spatial location of the cells, their developmental point in time as well as the disease state, the environmental exposure, and the lifestyle of the donor [4]. The success of such goals crucially depends on the development of technologies that enable multimodal and multiconditional measurements. At the forefront of these developments stands the field of spatially resolved transcriptomics (SRT), which has been widely recognized as one of the most promising biological technologies [5]. This review provides an overview of SRT technologies and discusses the technological and computational challenges of applying SRT in pathology.
Sequencing-based vs. Imaging-based Approaches
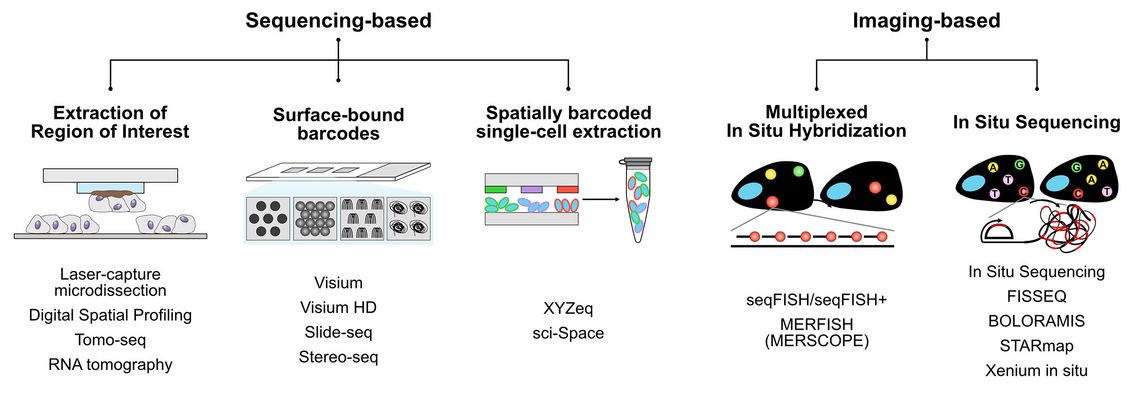
The SRT field can be largely divided into two methodological principles: Sequencing-based SRT and imaging-based SRT (Figure 1).
In sequencing-based SRT, different strategies are used to encode the position of an RNA molecule within a tissue section prior to extraction and quantification using next generation sequencing (NGS). One strategy is the extraction of regions of interest (ROIs) using methods such as laser-capture microdissection [6], Tomo-seq [7] or Digital Spatial Profiling (DSP) [8]. DSP has been commercialized in Nanostring’s GeoMX system and is now one of the most widely used SRT platforms. While the physical extraction of the ROIs allows flexibility in the downstream readouts, the resolution is limited, and the ROIs must to be known in advance. In 2016, Ståhl et al. introduced the method of Spatial Transcriptomics in a seminal paper [9]. This method, whose name is now used for the entire scientific field, uses unique surface-bound DNA barcodes, arrayed on a glass surface. After attaching a tissue section to the glass, molecular biological methods are used to label RNA molecules within the tissue section with these barcodes. NGS and subsequent computer analysis facilitate the mapping of sequencing reads to spatial locations. The Ståhl method has triggered the development of a series of novel technologies using different strategies to generate surface-bound DNA barcodes, including spots [10], beads [11, 12], clusters [13, 14], nanoballs [15], or microfluidic devices [16, 17]. Although improved manufacturing methods achieved ever smaller barcode features, diffusion effects set a natural limit to the resolution of such methods and prevent these methods from achieveing actual single-cell resolution. Methods such as XYZeq or sci-Space combine spatial barcoding with single-cell extraction methods to potentially overcome these limitations, but so far lacking in throughput and sensitivity [18, 19].
Unlike to sequencing-based SRT technologies, imaging-based methods are not affected by diffusion effects during the barcoding step and are therefore able to reaching sub-cellular resolution. A fundamental principle of all these methods is fluorescent in situ hybridization (FISH), in which fluorescently labelled oligonucleotides bind to their complementary target sequences, making them visible under a fluorescence microscope. However, the width of the fluorescence spectra allows the measurement of only 3-4 fluorochromes in parallel, which drastically limits the number of genes that can be characterized in parallel. The introduction of different combinatorial strategies enabled the detection of many more genes in parallel, and this development culminated in highly multiplexed methods such as seqFISH+ [20], or MERFISH [21], which is marketed as MERSCOPE platform by Viszgen. As the signal strength depends on the number of hybridized fluorescent probes, the application of FISH-based methods is limited in the case of short target sequences. One solution to this is rolling circle amplification (RCA), which allows the isothermal amplification of the target nucleic acids using so-called padlock probes and thus yields stronger signals and higher selectivity than common FISH methods. In In Situ Sequencing (ISS) RCA has been combined with sequential, image-based readouts of fluorescently labelled detection probes to identify target RNA molecules [22]. In recent years, a variety of different technologies such as FISSEQ [23], STARmap [24] or BOLORAMIS [25] have been developed, which used the general principle of in situ sequencing, allowing the quantification of 100s to 1000s of genes in tissue sections. Combining the strengths of both the original ISS method and FISSEQ, 10X Genomics commercialized in 2022 a novel method called Xenium In Situ [26, 27], which, among other things, was used to map the breast cancer tumor microenvironment [28]. Comparisons of the currently most widely used SRT platforms GeoMX, MERSCOPE, and Xenium in situ in recent benchmarking articles have revealed both the strengths and weaknesses of these methods [29 – 31].
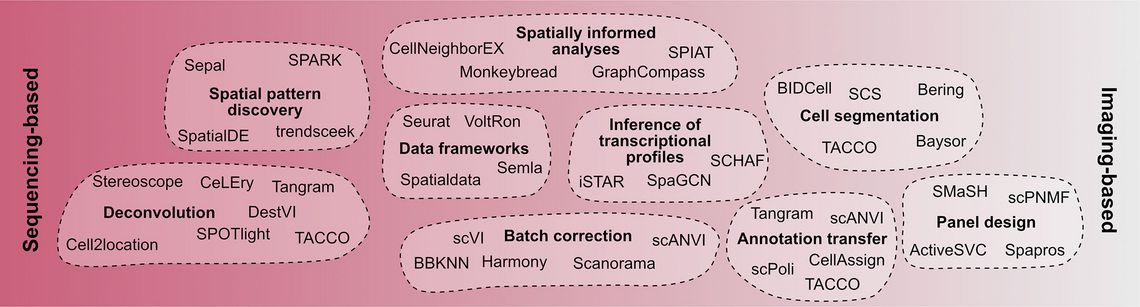
While early methods could only be applicable to fresh frozen samples, protocols have been developed to apply them also to FFPE samples, making the large pathological archives accessible for analysis. While this promises to revolutionize pathological research in the future, particularly the computational analysis of such datasets poses a number of challenges. These challenges and tools available to overcome them are discussed below (Figure 2).
Deconvolution of Spatial Transcriptomics Data
Sequencing-based methods such as Visium, Slide-seq or the recently published Visium HD technology, have resolutions from 50 µm down to 2 µm. Due to the technological design, each barcoded spot can contain transcripts from multiple cells, with the number of cells depending on the resolution of the method, the location of the spot relative to the tissue section, and the cell density within the tissue section. To infer cell type proportions as well as single-cell gene expression levels, computational deconvolution approaches have been developed. Most of these algorithms such as Stereoscope [32], cell2location [33], SPOTlight [34], DestVI [35], Tangram [36] or TACCO [37] use single-cell transcriptomics (scT) data as references to map single-cell information onto the SRT data. Other approaches like STdeconvolve use a reference-free approach and do not rely on scT datasets [38]. However, all these methods provide only computational approximations and are therefore not free from biases.
Cell Segmentation for Accurate Transcript Assignment
In contrast, imaging-based methods generate data consisting of the location of each measured transcript at a subcellular resolution, eliminating the need for data deconvolution. Instead, the accurate assignment of transcripts to cells becomes crucial, which brings cell segmentation into focus. In recent years, cell segmentation has been significantly improved using deep learning approaches in combination with multiplexed images, resulting in algorithms such as Cellpose [39,40] or Mesmer [41]. However, the accuracy of cell segmentation can vary between tissue types and alternative approaches such as Baysor [42], Bering [43] or BIDCell [44] exploit the transcript locations to augment the image data and improve cell segmentation. Although deconvolution is not required in imaging-based SRT methods, the integration of SRT data and scT data using tools such as TACCO [37] or Tangram [36], facilitates the transfer of cell type labels from annotated scT data to SRT data, thus combining the strengths of both technologies.
Optimizing Gene Panels for Targeted Imaging-Based Transcriptomics
So far, the most widely used imaging-based methods are targeted and rely on the prior determination of genes of interest in so-called gene panels. These gene panels determine which processes and cell types can be measured in the experiment and their design is therefore crucial. Various methods such as Spapros [45], SMaSH [46] or ActiveSVC [47] have been developed to derive optimal gene sets from scT datasets, making experiments more cost-effictive and improve transferability between scT and SRT experiments.
Addressing Batch Effects in Clinical Transcriptomic Data
Particularly in the context of clinical samples, the pre-processing steps and storage times can vary between samples, introducing batch effects in the resulting datasets which are independent of the methodology. Both for datasets from imaging-based and sequencing-based methods batch correction algorithms such as Harmony [48], Scanorama [49], or scVI [50] have been developed and compared in benchmarking studies [51].
Advancements in Predictive Modeling for Gene Expression
Unlike scT methods, SRT technologies combine image data and transcriptomic data. This allows researchers to train deep neural networks to predict the gene expression from histological images as demonstrated in SpaGCN [52], SCHAF [53] or iSTAR [54]. In the future, such algorithms could facilitate the inference of transcriptomic profiles and cell types based on histological stainings and thereby partially replace transcriptomic readouts and complement immunohistochemical stainings.
Enhancing Pathological Diagnostics
Current diagnostic workflows in pathology focus on the analysis of either histological images or sequencing-based readouts. However, SRT methods produce large data sets combining different modalities, which makes the analysis computationally resource-intensive and complex, demanding a bioinformatic skill set from (molecular) pathologists. In order to facilitate access to SRT methods to clinical researchers, integrated analytical frameworks that enable rapid visualization and exploration of data, while providing access to novel third-party analytical tools are important. The currently emerging frameworks, such as Seurat [55] and VoltRon [56] for R as well as Squidpy [57], SpatialData [58] and TissUUmaps [59] for Python, integrate the data modalities and offer a wide range of analyses, but require deeper bioinformatic knowledge, making integration into existing pathological frameworks difficult. The successful translation of knowledge from SRT experiments into improved treatments for patients will, however, rely on the integration of existing human expert knowledge with the rapidly evolving field of computational analyses. Therefore, the establishment and improvement of analytical frameworks are pivotal. Furthermore, for the correct interpretation of results and the integration of spatial transcriptomics methodologies into diagnostic workflows, training pathologists on these new data formats will be essential.
Conclusion
In conclusion, spatially resolved transcriptomics methods allow for the characterization of healthy and diseased tissue at an unprecedented depth and have the potential to revolutionize translational research. The currently observed increasing digitalization of pathological workflows [60] and the growing landscape of computational tools pose a challenge, but above all, present a unique opportunity to integrate these technologies into pathological research and improve the diagnosis and treatment of diseases.