Developing Robust AI Applications for Clinical Use: The Special Case of Pathology
DOI: https://doi.org/10.47184/tp.2024.01.04Robustness is a key requirement for any method in medicine, especially when the method in question is being used as part of a diagnostic process. This is particularly true for artificial intelligence-based decision support systems, which, although being used as a supportive tool, will ultimately influence diagnostic assessments. In pathology, attaining clinical robustness in AI methods poses a particularly challenging task, primarily due to the extensive diversity of digital images, which humans can adapt to far more easily. This paper presents factors that contribute to this challenge, but also identifies and evaluates common solutions to counteract domain shift, which is known to deteriorate the performance of artificial intelligence models.
Keywords: Clinical application, robustness, domain generalization
Introduction
Recent technological advances in artificial intelligence (AI) have made it possible to simplify workflows in ways that were previously unimaginable. AI excels at creating and understanding all types of human-generated content, such as text and images, paving the way for AI-based devices with high levels of automation. While AI-based medical decision support systems are currently being deployed on a large scale, with some solutions already FDA-cleared, the field of pathology has not yet moved significantly towards fully digital and computerized workflows. At first glance, it may seem surprising that we are now living in an age where we can reliably identify people from photographs or make vehicles drive at least partially autonomously, but in a field where the task is as specific and narrowly defined as the detection of a particular disease pattern or structural marker from a digital histopathology image, the solutions have yet to emerge. However, a closer look reveals that pathology is indeed a special case, as there is more than only one factor limiting this transition: Most pathology laboratories still rely heavily on analogue light microscopy for their diagnostic workflow, and digitising slides adds another step to the workflow that is not supported by current revenue models. In addition, solution providers have not widely offered AI algorithms that can be reliably integrated into workflows. Therefore, in the subsequent sections, let us take a closer look at why the development of robust algorithms is a particular challenge in pathology.
The path towards robust solutions
Deep neural networks, the predominant models utilized for image comprehension, acquire knowledge from data. Given a defined set of input data, the model learns during the training to reproduce the associated output data, e. g. objects in the image or a class label associated with the image. Nevertheless, the models do not learn how to process data that differs from the data they have encountered previously. This data is known as out-of-domain data. A transition in the image depiction, often termed as a domain shift concerning the training data, can stem from various factors in pathology. Pathology workflows lack standardization across different laboratories. Certainly, the origin of the tissue, i. e. the organ or even the type of tumor, impacts the morphological characteristics of the tissue. But even for the same tissue morphology, there is a long list of factors that affect the digital image, from the way it is fixed to other pre-analytical effects such as the thickness of the tissue sections and the staining protocol used. However, as recent research has shown [1], digitization equipment also contributes to significant visible changes in the image. In addition to the spectral characteristics of the light source and the image sensor, which alter the colour representation in the image, the optical paths also differ between microscopes/scanners, resulting in differences in depth of field and overall sharpness. All of this results in real (and unpredictable) drops in detection performance when evaluating even the same physical slides digitised by different devices (whole slide scanners or microscopes/cameras).
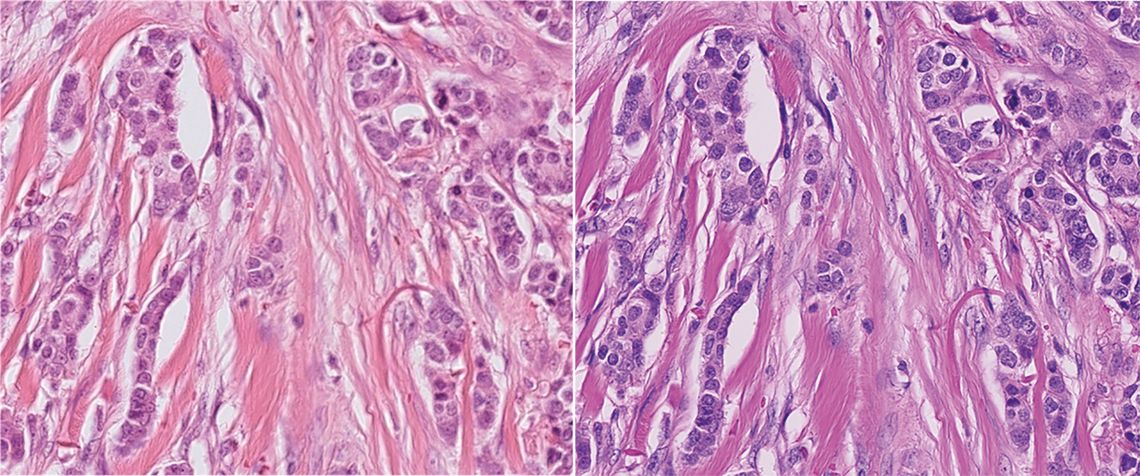
Figure 1: Sample of breast cancer scanned with two different scanners, showcasing the challenge for deep network robustness: Between the scanners, the depth of field, overall sharpness and especially color representation vary significantly, leading to a domain shift. Image taken from the MIDOG 2021 challenge [1].
Different strategies to counteract the domain shift have emerged over the last years [2], the most promising of which are data augmentation, domain alignment, and dataset enrichment. Data augmentation increases the diversity of the data by applying, e. g., geometric and colour transformations, to create new images representing the same content, and is now a widely used standard strategy in histopathology [1]. However, changing the image in this way and not altering the pathological information represented by the image is non-trivial and requires tuning of parameters, as color and shape serve as an important cue in histopathology, and, for some cases, might even deteriorate model performance [3]. Domain alignment follows the idea of making the model more robust by learning to ignore domain covariant factors, i. e. information within the image that varies with the domain but has no relevance to the task at hand. Domain alignment methods are typically applied by using a specific model architecture or loss function to reduce the domain dependency of the internal representations of the model. Although this is a very active area of research and many methods have achieved significant improvements in domain robustness, the much more effective way to achieve domain robustness is to expand the training datasets themselves.
Towards large-scale datasets
Considering the significant shift caused by the image capture device, we can at least reduce the cost of creating datasets by a significant factor: Glass slides can be digitised multiple times and registered with high accuracy using state-of-the-art algorithms [4], allowing direct translation of annotations between an initially annotated digital slide and re-scanned copies. However, even this data enhancement is not free: Re-scanned slides can have different degrees of sharpness, which can lead to a change in the diagnostically relevant information that may have been annotated on the original [5].
Another trend that is likely to transition from general image understanding to pathology is the use of textual descriptions for images. At least in institutions where the pathology diagnostic workflow is already fully digitised, the pathology reports, together with digital whole slide images, represent a valuable and largely free resource that we are only beginning to understand how to exploit to its full potential. Foundation models, trained on such large corpora, can make use of various modalities, and contribute to the models becoming more robust. However, as this is still a new area, its true impact will have to be assessed over the next few years.
Conclusion
Only a representative and sufficiently sized sample of the true data distribution, i. e., of digital images taken from routine diagnostic use cases in various clinical environments, can ultimately be used to create AI models that not only promise of being robust – but also are proven to be robust, because they have been validated on a representative and sufficiently large set of test cases. In this sense, pathology is no different from any other AI problem after all. Given that a large and well-annotated data corpus is available for a problem, a deep learning model can solve it. The success of these models in image and text understanding, or the recent success in generating high-fidelity images, is also rooted in this. We can thus expect to have solutions available that can support the diagnostic process in pathology within the next few years. However, the manner in which they will be utilized, and the possibility of enhancing workflow in pathology without potentially introducing diagnostic bias, are questions that initiate a new realm of research.

