AI-based image classification algorithms for Leukaemia diagnostics and hematologic cytomorphology: From single cells to molecular features
DOI: https://doi.org/10.47184/tp.2024.01.05Due to the progress of image analysis and classification systems in recent years, algorithms have been developed that support morphologic examination of both single cells and tissue samples. These algorithms are typically developed using data-driven strategies, which require comprehensive, large-scale datasets.In the diagnostic workup of hematopoietic malignancies, cytomorphologic examination and differentiation represents a key first step. In recent years, the availability of large-scale, high-quality datasets of single leukocytes from peripheral blood and bone marrow has led to the development of diagnostic support algorithms for this modality. These methods not only allow a faster and more consistent classification of diagnostically relevant cell types, but also pave the way for integrated analysis of cytomorphologic and molecular findings.
Keywords: Leukaemia diagnostics, cytomorphology, explainable AI
Diagnostic background
Morphologic examination of bone marrow and peripheral blood represents a time-honoured diagnostic method developed in the 19th century [1] which is fast, low-cost and widely available [2]. It has therefore maintained its role in the evaluation of hematologic malignancies, among an increasing number of molecular tests that are often necessary to fully characterise hematologic malignancies [3]. For morphologic examination, samples are typically processed using a standard panoptic stain, e. g. according to the protocols developed by Pappenheim or Wright [4]. Evaluation is then performed by trained human examiners using a high-resolution light microscope. This process involves the differentiation of several hundred cells according to a scheme of about 20 morphologic cell types [5], which can be tedious, time-consuming, and prone to substantial inter- and intra-rater variabilities [6,7].
Hence, there is a diagnostic need for developing decision support algorithms that can quickly analyse large numbers of leukocytes, and evaluate their morphologic distributions, rendering the process faster and more reproducible.
Large datasets: A key requirement
While the overall tissue architecture is a key factor in the histologic evaluation of solid tumors, cytomorphology focuses on the properties of single cells. In the context of leukaemia diagnostics, typically at least 200 leukocytes are evaluated to obtain a representative distribution of cell types [8]. Hence, most cytomorphology datasets consist of annotated single-cell images rather than scans of an entire smear sample. Leukocyte subtype differentiation requires the ability to distinguish subcellular structures such as cytoplasmic granulation or features of nuclear structure. Hence, sample digitization must be performed at a sufficiently high overall resolution, which is typically achieved using at least 40x objective magnification. For analysis in the context of supervised AI-based algorithm development, datasets also require gold standard labelling by trained examiners. As is often the case in medical data analysis, an insufficient number of available datasets from a diverse range of sources can hamper progress in algorithm development and make multicentre evaluation difficult. To improve this situation, a series of single-cell image cell datasets have recently been published, containing samples of peripheral blood [9–13] and bone marrow smears [14, 15]. These datasets can either be focused on specific entities such as Acute Myeoloid Leukaemia (AML) [11, 12] or broader classes of hematopoietic malignancies and non-neoplastic alterations [14]. Available datasets can be annotated either at the level of individual morphological cell types or using an overall, case-level diagnostic label. Datasets covering different disease entities and possessing different annotation depths lend themselves to various computational analysis technologies.
Single-cell and collective classification
Approaching cytomorphologic evaluation as an image classification task on single leukocytes, many recent advances in deep learning-based image classification can be used to train algorithms that attain a high accuracy in clinically relevant diagnostic questions. Successful applications of image deep neural networks include the detection of blast cells in the peripheral blood of patients diagnosed with acute myeloid leukaemia [16], and the classification of single leukocytes from bone marrow samples [17], both of which were developed using the ResNeXt network structure [18]. In some subtasks such as blast detection, algorithms can attain the classification performance of human examiners [16]. As in many image classification tasks, neural networks outperform algorithms relying on hand-crafted feature extraction. However, employing a supervised training strategy on a single leukocyte level relies on a large number of gold-standard annotations provided by human investigators, which can be difficult and time-consuming to obtain and exhibit significant inter-rater variability. Methods such as Multiple Instance Learning (MIL) aim to reduce the number of expert annotations needed and predict collective features of many single cells from a “bag” of single-cell images [19]. For example, this method can be employed to train a prediction algorithm that infers specific molecular alterations from a set of single-cell images in peripheral blood smears from patients diagnosed with acute myeloid leukaemia [9, 20] or myelodysplastic syndrome (MDS) [9]. This strategy can detect rare diagnostic cells within the smear, which can then be examined and checked for specific, human-interpretable features, thus allowing verification of the prediction and pointing towards novel, hitherto unknown morphologic features correlated with molecular alterations.
Levels of explainability and interpretability
In a diagnostic setting, classification decisions are typically required to offer a degree of explainability in order to render predictions transparent to human supervisors and allow for error detection. Numerous approaches have been developed to extract explanations for deep-learning-based image classification algorithms, which highlight key regions within an image according to their importance for the final classification decision [21]. In the context of deep multiple-instance learning, an attention score can be attributed to single-cell images, which allows ranking single leukocytes according to their importance for the overall, bag-level classification (Fig. 1B).
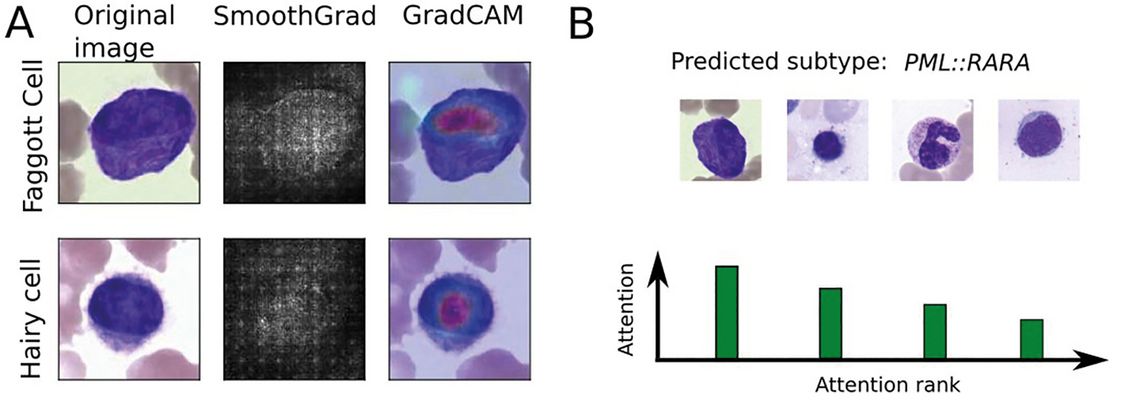
Figure 1: Levels of explainability: (A) Two common explainability methods, SmoothGrad [25] and GradCAM [25], highlighting the most important areas in two correctly classified single-cell images from bone marrow samples. (B) Schematic depiction of attention distribution obtained from a multiple instance learning algorithm, highlighting single cells with a high contribution to the final, patient-level diagnosis.
Both methodologies can also be combined [22]. Importantly, explainability methods cannot demonstrate the correctness of a specific classification. Instead, they can highlight errors such as high attention to background or confounding content [23].
Robustness and generalisability
In many use cases, machine learning classifiers need to be robust and generalisable to be practically usable as diagnostic decision support. A key measure of generalizability is performance on a large and diverse cohort of cases from several independent sites with potentially distinct preanalytic conditions, obtained by a range of different equipment. Since sharing data from diagnostic processes is often only possible in the limited context of research studies, and local datasets may be smaller compared to large training cohorts, methods have been devised to adapt trained classifiers to novel, yet unseen domains [24]. The ultimate measure of the reliability of a cytomorphologic classification algorithm is its local validation, which is a key requirement for these algorithms to be used productively.
Conclusion
Classifiers for the evaluation of cytological samples from leukaemia patients have recently shown significant improvements. Using machine learning-based approaches and large, high-quality datasets, powerful classifiers can be developed for both single-cell classification and collective classification of a large set of individual leukocytes. These classifiers can be analysed using explainable AI methods, allowing tests for confounding information contained in the training images, and helping to elucidate morpho-molecular relationships. Current algorithms promise to integrate several different diagnostic modalities, such as imaging, clinical data or histologic samples, in the future, making AI-based evaluation even more comprehensive.

