Deep-learning-based interpretability and the ExaMode project in histopathology image analysis
DOI: https://doi.org/10.47184/tp.2023.01.05With digital clinical workflows in histopathology departments, the possibility to use machine-learning-based decision support is increasing. Still, there are many challenges despite often good results on retrospective data. Explainable AI can help to find bias in data and also integrated decision support with other available clinical data. The ExaMode project has implemented many tools and automatic pipelines for such decision support. Most of the algorithms are available for research use and can thus be of help for other researchers in the domain.
Keywords: Computational pathology, decision support, machine learning, explainability
Introduction
The ExaMode1 project is a research project funded by the European Union’s Horizon 2020 framework program (running from 2019–2023). Seven complementary European research partners work on reaching the main project objectives, notably two Universities, two medical centers, two companies and a national supercomputing center.
The goals of the project are linked to very-large-scale data analysis, for which histopathology is an almost perfect use case with extremely large images and very large numbers being produced in many hospitals. The very-large-scale analysis aims to build data-driven decision support tools for clinicians. A challenge in image-based decision support is the manual annotation of images to train deep learning models, which is extremely expensive and has some subjectivity. For this reason, the project aims to use weak annotations [2, 3], so slide level labels for staging/grading/etc. instead of strong annotations, so annotated pixels. All data of a case, such as text reports and images are combined for multimodal data analysis. Text is mapped onto semantics for a better understanding, and domain specific ontologies were developed for the project use cases [4]. Multimodal learning aims to gain from the text and semantics and learn decision support on the images [1]. This has the advantage that learning does not require any human intervention if sufficiently large data sets are available.
Project structure
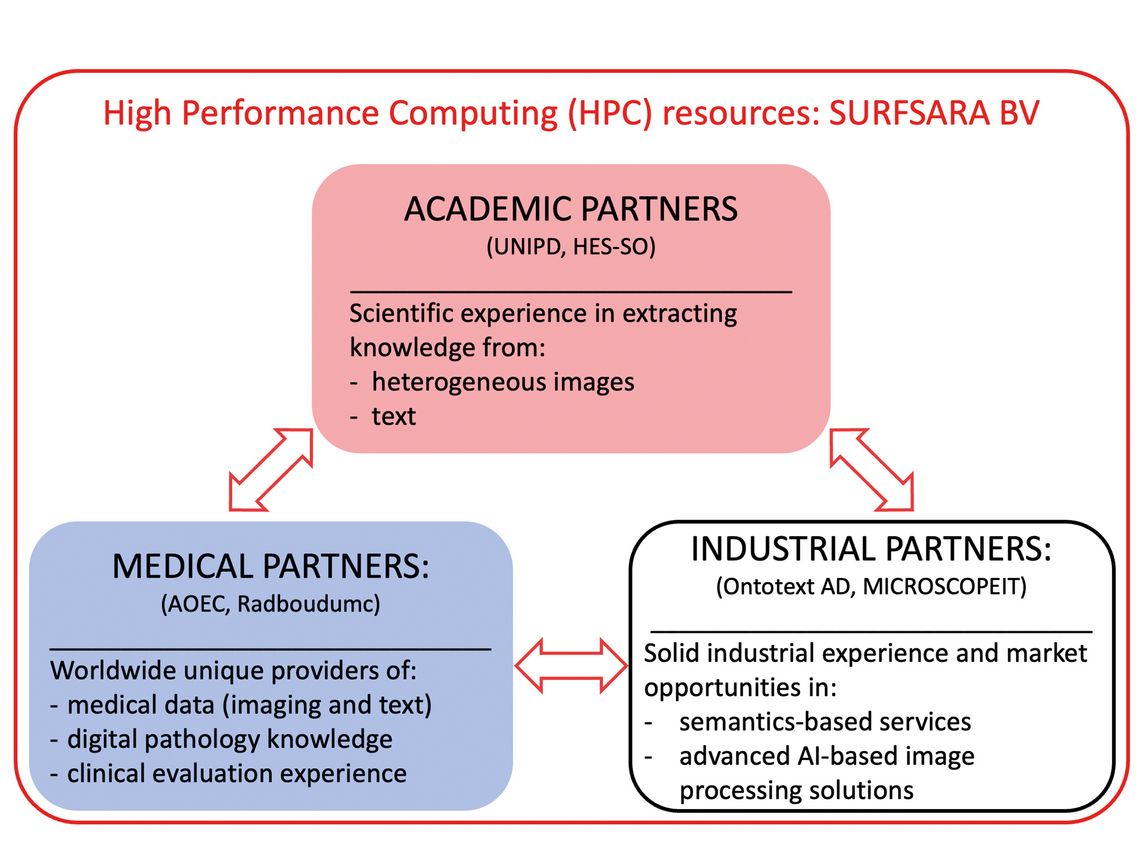
The entire physical computing infrastructure (storage and CPU/GPU computing) of the project is at SURFSARA, the Dutch national supercomputing center, which allows to develop scalable approaches for thousands of histopathology images (of around 10 GB each). Safe storage and secure computing with strict access control are also guaranteed with this. Figure 1 details the partners of the project.
Two academic partners assure the research on machine learning for text data and for imaging data. The two companies have a similar focus, so one for text analysis/semantics and one for the image analysis. The two clinical partners are also complementary, with Radboud Medical Center assuring practical implementation and data acquisition and the Catania hospital the data acquisition, annotation and tests in clinical settings.
Main project results
The project started in developing a solid foundation for the work to be done by requesting ethics and collecting data from public resources and in the partner hospitals. Use cases (lung cancer, colon cancer, uterus/cervix cancer and celiac disease) were chosen and then semantic ontologies were created for each use cases based on the pathology reports of each of the covered areas [4]. This allows to map reports in several languages (Dutch, Italian, English) onto concepts that can then be used as weak labels for the machine learning.
An image viewer was developed by one of the commercial partners as a vehicle for annotating the whole slide images and also for the decision support tools.
The image analysis work started with some challenges such as stain heterogeneity that can limit results in classification tasks of the images. We compared several stain normalization and data augmentation approaches and then developed a novel data augmentation process that learns the types of augmentation that actually are present in the specific image collection [5], leading to much better results than other approaches.
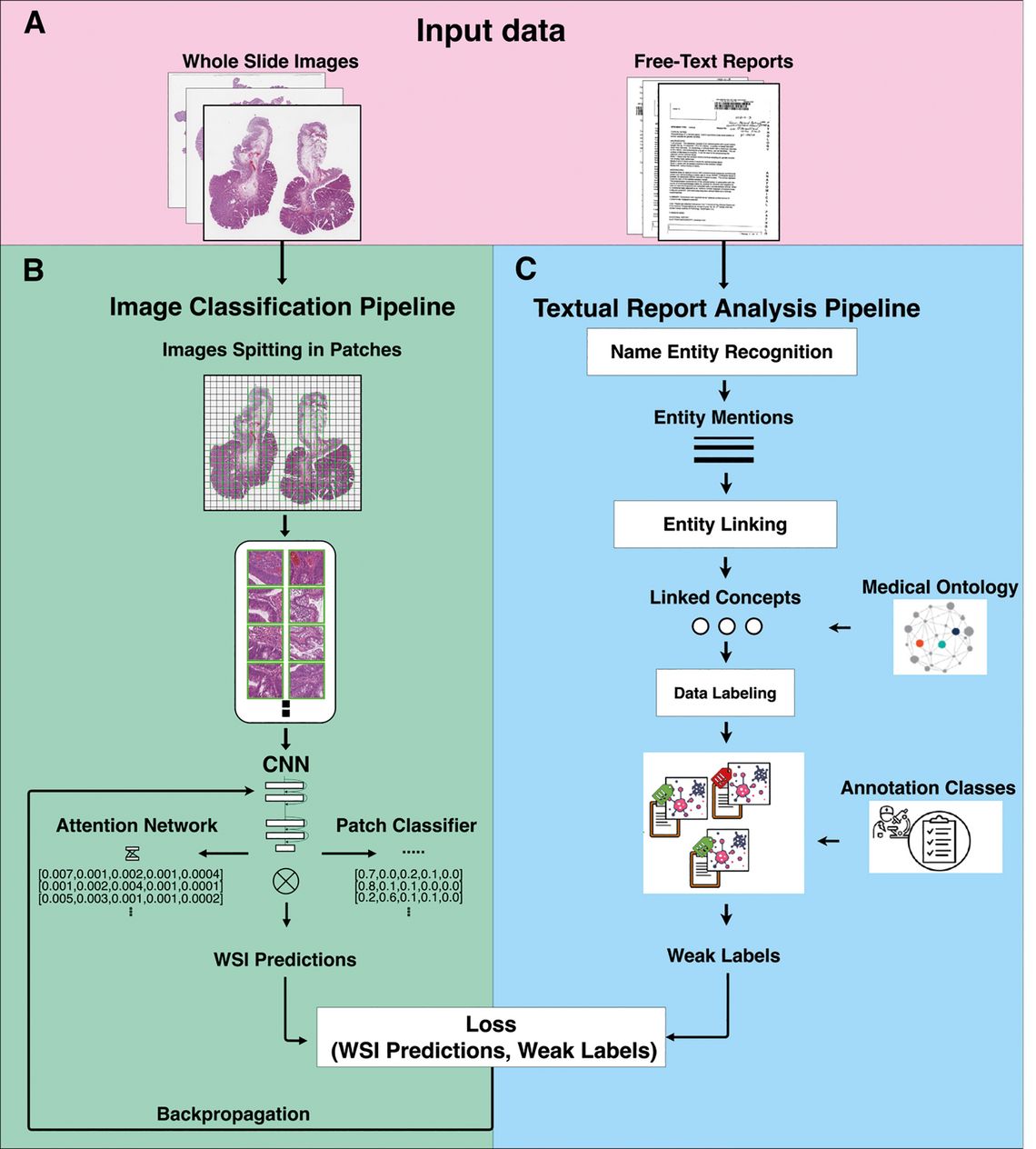
A major part of the work focused on using global labels for the machine learning aspects, which reduces the manual annotation effort massively [2, 3] by combining few strong annotations with many weak labels. Finally, multimodal learning [1] allows to bypass the manual annotation task completely by using labels automatically extracted from the text reports and learning with these. Figure 2 details the workflow for this approach.
One challenge when integrating image-based decision support based on deep learning is that deep learning is per se a black box and algorithmic decisions are hard to be explained. Thus, explainable deep learning approaches are an important part for system integration. We started in developing first a taxonomy for explainable AI, as many different terms are used with sometimes contradictory meanings [8] and varying aspects of explainability/ interpretability.
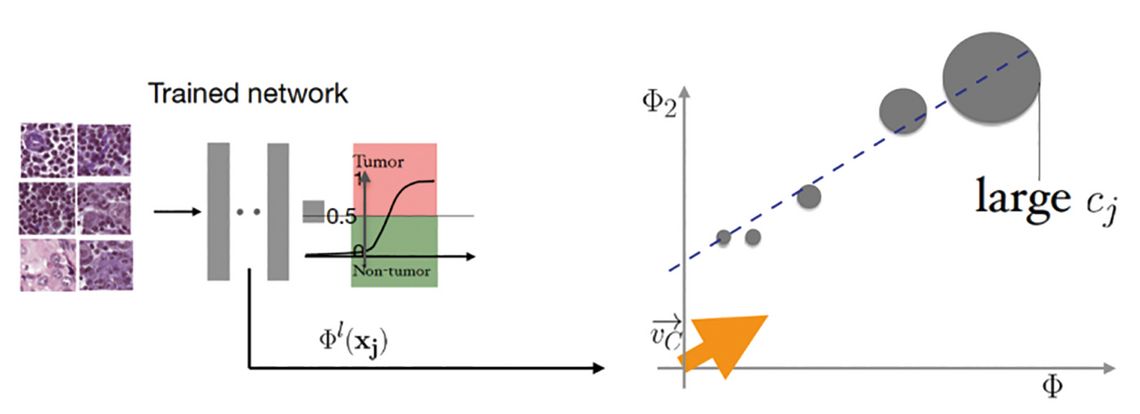
We developed several approaches for explanations from heat maps of image regions responsible for decisions and their adaptions to histopathology images towards regression concept vectors (illustrated in Figure 3) that allow to map the internal decision-making process of a deep neural network post-hoc onto human understandable features that we defined in collaboration with clinicians (such as size of the nuclei, nuclei border heterogeneity or internal nuclei heterogeneity in texture) [6, 7].
The advantage is that the decision can be explained with features understandable to humans but these need to be manually defined and thus can be biased. Work is currently ongoing for finding such concepts automatically by data analysis, so go beyond what has been described by humans.
Conclusions
Decision support in digital pathology still has many challenges ahead but technology clearly has advanced strongly. Stain normalization is now less of a problem, with many public data sets allowing to adapt also important variations in staining and with learned data augmentation adding efficiency. Manual annotations of regions can well be limited when using weak supervision for the learning or even unsupervised learning. Particularly learning labels from reports in a fully automatic way seems to be the future of digital pathology when image data with reports are available. A full integration of these tools into clinical viewers is required, as well as tools that allow to interpret the decisions of algorithms and to integrate this information with other data on a specific case. User studies are really required to learning about other, individual factors concerning decision support or possible also fully automatic approaches for triage of cases or in screening tasks.